RegulonDB
HipAB matrix and aligment
matrix-quality result
Command: matrix-quality -v 1 -ms ./data/Matrices_NR/HipAB/HipAB.EcolK12_1nt_upstream.29.meme
Figures
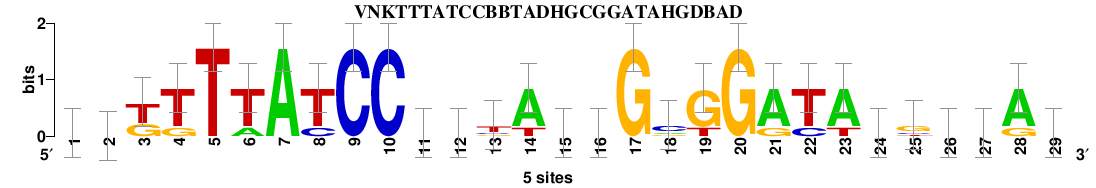
Matrix logo
Decreasing cumulative distributions (dCDF)
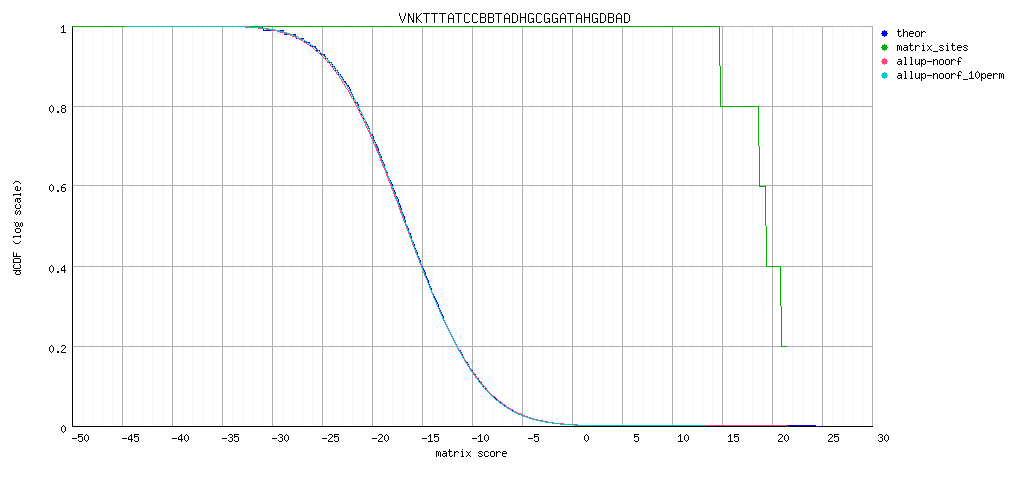
Decreasing cumulative distributions (dCDF), logarithmic Y axis

ROC curve (logarithmic X axis)
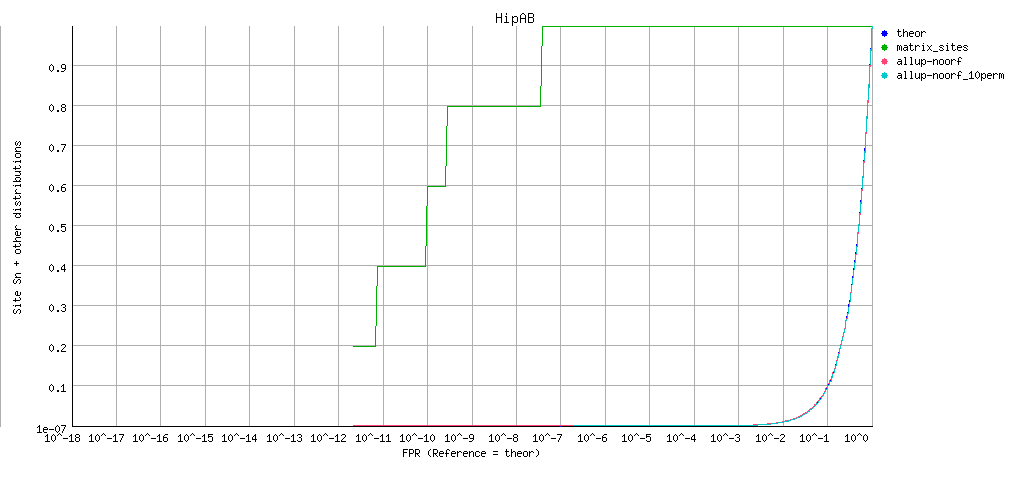
Matrix information
; convert-matrix -v 1 -from transfac -i /HipAB.EcolK12_1nt_upstream.29.meme_quality_logo
; Input files
; input /HipAB.EcolK12_1nt_upstream.29.meme_quality_matrix.tf
; prior /HipAB.EcolK12_1nt_upstream.29.meme_quality1nt_upstream-noorf_Escherichia_coli_GCF_000005845.2_ASM584v2-ovlp-1str.freq.gz_inclusive.tab
; Input format transfac
; Output files
; output /HipAB.EcolK12_1nt_upstream.29.meme_quality_matrix_info.txt
; Output format tab
; pseudo-weight 1
; Background model
; Bernoulli model (order=0)
; Strand undef
; Background pseudo-frequency 0.01
; Residue probabilities
; a 0.29093
; c 0.20756
; g 0.20418
; t 0.29732
A 2 1 0 0 0 1 5 0 0 0 0 0 0 4 2 2 0 1 0 0 4 0 4 2 0 2 0 4 2
C 2 2 0 0 0 0 0 1 5 5 1 2 1 0 0 2 0 3 0 0 0 1 0 1 1 0 2 0 0
G 1 1 2 1 0 0 0 0 0 0 2 2 1 0 2 0 5 1 4 5 1 0 0 0 3 2 1 1 1
T 0 1 3 4 5 4 0 4 0 0 2 1 3 1 1 1 0 0 1 0 0 4 1 2 1 1 2 0 2
//
A 0.4 0.2 0.0 0.0 0.0 0.2 0.9 0.0 0.0 0.0 0.0 0.0 0.0 0.7 0.4 0.4 0.0 0.2 0.0 0.0 0.7 0.0 0.7 0.4 0.0 0.4 0.0 0.7 0.4
C 0.4 0.4 0.0 0.0 0.0 0.0 0.0 0.2 0.9 0.9 0.2 0.4 0.2 0.0 0.0 0.4 0.0 0.5 0.0 0.0 0.0 0.2 0.0 0.2 0.2 0.0 0.4 0.0 0.0
G 0.2 0.2 0.4 0.2 0.0 0.0 0.0 0.0 0.0 0.0 0.4 0.4 0.2 0.0 0.4 0.0 0.9 0.2 0.7 0.9 0.2 0.0 0.0 0.0 0.5 0.4 0.2 0.2 0.2
T 0.0 0.2 0.5 0.7 0.9 0.7 0.0 0.7 0.0 0.0 0.4 0.2 0.5 0.2 0.2 0.2 0.0 0.0 0.2 0.0 0.0 0.7 0.2 0.4 0.2 0.2 0.4 0.0 0.4
//
A 0.3 -0.3 -1.8 -1.8 -1.8 -0.3 1.1 -1.8 -1.8 -1.8 -1.8 -1.8 -1.8 0.9 0.3 0.3 -1.8 -0.3 -1.8 -1.8 0.9 -1.8 0.9 0.3 -1.8 0.3 -1.8 0.9 0.3
C 0.6 0.6 -1.8 -1.8 -1.8 -1.8 -1.8 -0.0 1.4 1.4 -0.0 0.6 -0.0 -1.8 -1.8 0.6 -1.8 0.9 -1.8 -1.8 -1.8 -0.0 -1.8 -0.0 -0.0 -1.8 0.6 -1.8 -1.8
G -0.0 -0.0 0.6 -0.0 -1.8 -1.8 -1.8 -1.8 -1.8 -1.8 0.6 0.6 -0.0 -1.8 0.6 -1.8 1.4 -0.0 1.2 1.4 -0.0 -1.8 -1.8 -1.8 1.0 0.6 -0.0 -0.0 -0.0
T -1.8 -0.3 0.6 0.9 1.1 0.9 -1.8 0.9 -1.8 -1.8 0.3 -0.3 0.6 -0.3 -0.3 -0.3 -1.8 -1.8 -0.3 -1.8 -1.8 0.9 -0.3 0.3 -0.3 -0.3 0.3 -1.8 0.3
//
A 0.1 -0.1 -0.1 -0.1 -0.1 -0.1 1.0 -0.1 -0.1 -0.1 -0.1 -0.1 -0.1 0.6 0.1 0.1 -0.1 -0.1 -0.1 -0.1 0.6 -0.1 0.6 0.1 -0.1 0.1 -0.1 0.6 0.1
C 0.2 0.2 -0.1 -0.1 -0.1 -0.1 -0.1 -0.0 1.2 1.2 -0.0 0.2 -0.0 -0.1 -0.1 0.2 -0.1 0.5 -0.1 -0.1 -0.1 -0.0 -0.1 -0.0 -0.0 -0.1 0.2 -0.1 -0.1
G -0.0 -0.0 0.2 -0.0 -0.1 -0.1 -0.1 -0.1 -0.1 -0.1 0.2 0.2 -0.0 -0.1 0.2 -0.1 1.3 -0.0 0.9 1.3 -0.0 -0.1 -0.1 -0.1 0.5 0.2 -0.0 -0.0 -0.0
T -0.1 -0.1 0.3 0.6 1.0 0.6 -0.1 0.6 -0.1 -0.1 0.1 -0.1 0.3 -0.1 -0.1 -0.1 -0.1 -0.1 -0.1 -0.1 -0.1 0.6 -0.1 0.1 -0.1 -0.1 0.1 -0.1 0.1
//
; Sites 5
>site_0
GCGTTTATCCCGTAGAGCGGATAAGATGT
>site_1
CTTTTTATCCGCGATCGCGGATATCGCAG
>site_2
AGTTTTATCCTTTAGTGAGGATAAGTCAA
>site_3
AAGTTTATCCGCTTAAGGGGATATTATAA
>site_4
CCTGTAACCCTGCAACGCTGGCTCGGGAT
;
; Matrix parameters
; Number of sites 5
; Columns 29
; Rows 4
; Alphabet A|C|G|T
; Prior A:0.290929484294843|C:0.207560750607506|G:0.204184816848168|T:0.297324948249482|a:0.290929484294843|c:0.207560750607506|g:0.204184816848168|t:0.297324948249482
; program transfac
; matrix.nb 1
; accession VNKTTTATCCBBTADHGCGGATAHGDBAD
; AC VNKTTTATCCBBTADHGCGGATAHGDBAD
; id VNKTTTATCCBBTADHGCGGATAHGDBAD
; name VNKTTTATCCBBTADHGCGGATAHGDBAD
; description mcktTtAtCCkstarmGcGGatawgryaw
; statistical_basis 5 sequences
; sites 5
; nb_sites 5
; min.prior 0.204185
; alphabet.size 4
; max.bits 2
; total.information 13.1378
; information.per.column 0.453027
; max.possible.info.per.col 1.58873
; consensus.strict ccttTtAtCCggtagcGcGGataaggcaa
; consensus.strict.rc TTGCCTTATCCGCGCTACCGGATAAAAGG
; consensus.IUPAC mcktTtAtCCkstarmGcGGatawgryaw
; consensus.IUPAC.rc WTRYCWTATCCGCKYTASMGGATAAAMGK
; consensus.regexp [ac]c[gt]tTtAtCC[gt][cg]ta[ag][ac]GcGGata[at]g[ag][ct]a[at]
; consensus.regexp.rc [AT]T[AG][CT]C[AT]TATCCGC[GT][CT]TA[CG][AC]GGATAAA[AC]G[GT]
; residues.content.crude.freq a:0.2483|c:0.2000|g:0.2483|t:0.3034
; G+C.content.crude.freq 0.448276
; residues.content.corrected.freq a:0.2554|c:0.2013|g:0.2409|t:0.3024
; G+C.content.corrected.freq 0.442187
; min(P(S|M)) 2.29836e-41
; max(P(S|M)) 1.13458e-07
; proba_range 1.13458e-07
; Wmin -50.5
; Wmax 24.7
; Wrange 75.2
; logo file:/HipAB.EcolK12_1nt_upstream.29.meme_quality_logo_m1.png
; logo file:/HipAB.EcolK12_1nt_upstream.29.meme_quality_logo_m1_rc.png
;
; Host name sinik
; Job started 2019-06-03.160931
; Job done 2019-06-03.160931
; Seconds 0.59
; user 0.59
; system 0.07
; cuser 0.44
; csystem 0.05




