RegulonDB
IHF matrix and aligment
matrix-quality result
Command: matrix-quality -v 1 -ms ./data/Matrices_NR/IHF/IHF.EcolK12_2nt_upstream.14.meme
Figures
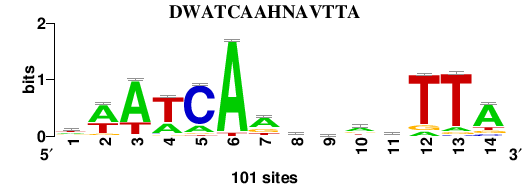
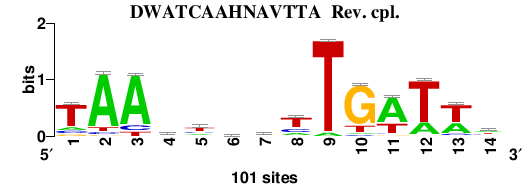
Matrix logo
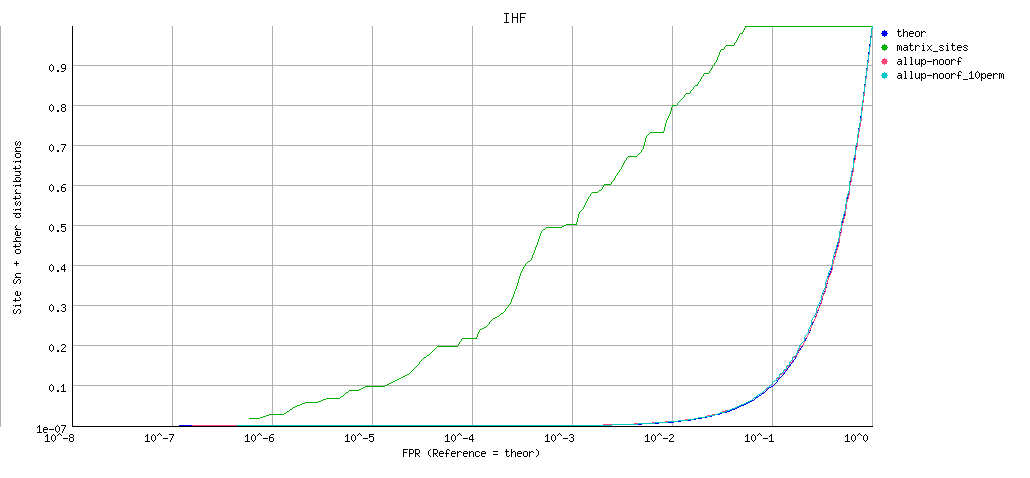
Decreasing cumulative distributions (dCDF)

Decreasing cumulative distributions (dCDF), logarithmic Y axis

ROC curve (logarithmic X axis)
Matrix information
; convert-matrix -v 1 -from transfac -i /IHF.EcolK12_2nt_upstream.14.meme_quality_logo
; Input files
; input /IHF.EcolK12_2nt_upstream.14.meme_quality_matrix.tf
; prior /IHF.EcolK12_2nt_upstream.14.meme_quality2nt_upstream-noorf_Escherichia_coli_GCF_000005845.2_ASM584v2-ovlp-1str.freq.gz_inclusive.tab
; Input format transfac
; Output files
; output /IHF.EcolK12_2nt_upstream.14.meme_quality_matrix_info.txt
; Output format tab
; pseudo-weight 1
; Background model
; Strand undef
; Background pseudo-frequency 0.01
; Residue probabilities
; a 0.29114
; c 0.20781
; g 0.20402
; t 0.29702
A 33 56 75 27 15 96 55 24 34 49 29 9 8 69
C 9 2 1 4 78 0 6 23 22 13 21 0 3 9
G 24 8 2 4 4 0 23 16 21 20 35 10 5 11
T 35 35 23 66 4 5 17 38 24 19 16 82 85 12
//
A 0.3 0.6 0.7 0.3 0.1 0.9 0.5 0.2 0.3 0.5 0.3 0.1 0.1 0.7
C 0.1 0.0 0.0 0.0 0.8 0.0 0.1 0.2 0.2 0.1 0.2 0.0 0.0 0.1
G 0.2 0.1 0.0 0.0 0.0 0.0 0.2 0.2 0.2 0.2 0.3 0.1 0.1 0.1
T 0.3 0.3 0.2 0.6 0.0 0.1 0.2 0.4 0.2 0.2 0.2 0.8 0.8 0.1
//
A 0.1 0.6 0.9 -0.1 -0.7 1.2 0.6 -0.2 0.1 0.5 -0.0 -1.2 -1.3 0.8
C -0.8 -2.3 -2.9 -1.6 1.3 -4.6 -1.2 0.1 0.0 -0.5 0.0 -4.6 -1.9 -0.8
G 0.2 -0.9 -2.2 -1.6 -1.6 -4.6 0.1 -0.3 0.0 -0.0 0.5 -0.7 -1.4 -0.6
T 0.2 0.2 -0.3 0.8 -2.0 -1.7 -0.6 0.2 -0.2 -0.5 -0.6 1.0 1.0 -0.9
//
A 0.0 0.4 0.7 -0.0 -0.1 1.1 0.3 -0.0 0.0 0.2 -0.0 -0.1 -0.1 0.6
C -0.1 -0.0 -0.0 -0.1 1.0 -0.0 -0.1 0.0 0.0 -0.1 0.0 -0.0 -0.1 -0.1
G 0.0 -0.1 -0.0 -0.1 -0.1 -0.0 0.0 -0.0 0.0 -0.0 0.2 -0.1 -0.1 -0.1
T 0.1 0.1 -0.1 0.5 -0.1 -0.1 -0.1 0.1 -0.1 -0.1 -0.1 0.8 0.9 -0.1
//
; Sites 101
>site_0
AAATCAATGAGTTA
>site_1
TTATCAATAAGTTA
>site_2
AAATCAATAACTTA
>site_3
GAATCAATGAATTA
>site_4
TTATCAACAAGTTA
>site_5
ATATCAAACAGTTA
>site_6
AAATCAATAGATTA
>site_7
GATTCAATGAGTTA
>site_8
AAATCAAGGAATTA
>site_9
TAATCAGTACGTTA
>site_10
ATATCAATGGCTTA
>site_11
TATTCAATTAGTTA
>site_12
AAAACAATGAATTA
>site_13
AAAACAAATAGTTA
>site_14
ATAACAATAAATTA
>site_15
TAATCAAATATTTA
>site_16
GAATCAGGCTGTTA
>site_17
TTAACAACAGGTTA
>site_18
AAATCATAAACTTA
>site_19
TTATCAGCGTGTTA
>site_20
AAAACAACAATTTA
>site_21
ATATCATTTAATTA
>site_22
AAATCAACTAATTC
>site_23
AAATCAAAAAGATA
>site_24
AAATCAAAATGTTG
>site_25
AAATCAGGTAGTTG
>site_26
GATTCAGCACCTTA
>site_27
AATTCAATAAATTG
>site_28
TAATCAAGCACTAA
>site_29
ATTTCAGTCATTTA
>site_30
AAAACACTCAATTA
>site_31
AAATCTATAAATTA
>site_32
AGAACAATTGGTTA
>site_33
ATATAAATAGATTA
>site_34
TAATCAGACGCTTG
>site_35
AAATAAATCTATTA
>site_36
TTATCAATGCCGTA
>site_37
ATATCAGACAATTC
>site_38
GTAACAACAGGGTA
>site_39
TAATCATCTAATTT
>site_40
GAACCAACTGCTTA
>site_41
GAATCAACGAGATG
>site_42
CAAACAATGCTTTA
>site_43
TAATCAACACGTCA
>site_44
TTTACAGTCTGTTA
>site_45
GTTACATTTAGTTA
>site_46
TAATCACTGTGTTG
>site_47
TTATTAGTAAGTTA
>site_48
CTATCAATATATTC
>site_49
GAATTAAAAAATTA
>site_50
TATTCAGCGTGTTC
>site_51
GAATCAATAATGTT
>site_52
CTAACAATGAGATA
>site_53
GCATCAACGCCTTA
>site_54
TTTACATGCACTTA
>site_55
GAGTCAGAGAATTA
>site_56
GTTACATCAATTTA
>site_57
TAATAAATCTATTG
>site_58
TTAACAAAAGTTTC
>site_59
AAACCAAATCTTTA
>site_60
TTTTCAAATTATTC
>site_61
TTATTAGCCGCTTA
>site_62
AAACCATACCCTTA
>site_63
TTATAAAGGGCTTT
>site_64
TTGACAAAAGGTTA
>site_65
GAACCAAATAGTAA
>site_66
TTCTCAGTATGTTA
>site_67
TCATCACTTTGTTA
>site_68
AGATCACTTAATTT
>site_69
CGAACAAGGACTTT
>site_70
CTATCACAGGATTG
>site_71
GAATGAATAGTTTT
>site_72
GAAGCACGCAGGTA
>site_73
AGATCAAGCCTTAA
>site_74
ATTTAAGGAACTTT
>site_75
TTTTCATTTCAATA
>site_76
TTTAAAGGTATTTA
>site_77
TAATCAAGTTTTGT
>site_78
GAAGCATCCCGTGA
>site_79
CATTCATGTTATTC
>site_80
CAATCAGCCTGTCT
>site_81
GAATGAGATTATTG
>site_82
CTTACATTCGGATA
>site_83
TGAAAAACAAATGA
>site_84
AATTCTGCAATGTA
>site_85
TAAAAATCATCATA
>site_86
TTTGAAAACCCTTA
>site_87
TGAAAAATCTCATA
>site_88
GAATATAAAATTTT
>site_89
GTTTAATATGCTTT
>site_90
GATTGATGAATTTC
>site_91
GGATCAGGCTTATG
>site_92
TATTTATAGAGTAA
>site_93
ATAGCAACTGCGGA
>site_94
AAATGTGCAAATTC
>site_95
TTAACAGTTGCGAT
>site_96
TATACAAATCAGCA
>site_97
CGAAAAAGCGATAA
>site_98
AATTCTTTGAGGAA
>site_99
GAAAAAGAGGAAGA
>site_100
GAAAAATCAAGGAG
;
; Matrix parameters
; Number of sites 101
; Columns 14
; Rows 4
; Alphabet A|C|G|T
; Prior A:0.291144263562772|C:0.207810681831126|G:0.204024081631615|T:0.297020972974487|a:0.291144263562772|c:0.207810681831126|g:0.204024081631615|t:0.297020972974487
; program transfac
; matrix.nb 1
; accession DWATCAAHNAVTTA
; AC DWATCAAHNAVTTA
; id DWATCAAHNAVTTA
; name DWATCAAHNAVTTA
; description dwatCAryvastTa
; statistical_basis 101 sequences
; sites 101
; nb_sites 101
; min.prior 0.204024
; alphabet.size 4
; max.bits 2
; total.information 4.95417
; information.per.column 0.353869
; max.possible.info.per.col 1.58952
; consensus.strict taatCAataagtTa
; consensus.strict.rc TAACTTATTGATTA
; consensus.IUPAC dwatCAryvastTa
; consensus.IUPAC.rc TAASTBRYTGATWH
; consensus.regexp [agt][at]atCA[ag][ct][acg]a[cg]tTa
; consensus.regexp.rc TAA[CG]T[CGT][AG][CT]TGAT[AT][ACT]
; residues.content.crude.freq a:0.4095|c:0.1351|g:0.1294|t:0.3260
; G+C.content.crude.freq 0.264498
; residues.content.corrected.freq a:0.4083|c:0.1358|g:0.1302|t:0.3257
; G+C.content.corrected.freq 0.265942
; min(P(S|M)) 1.89094e-20
; max(P(S|M)) 0.000346904
; proba_range 0.000346904
; Wmin -24.4
; Wmax 10
; Wrange 34.4
; logo file:/IHF.EcolK12_2nt_upstream.14.meme_quality_logo_m1.png
; logo file:/IHF.EcolK12_2nt_upstream.14.meme_quality_logo_m1_rc.png
;
; Host name sinik
; Job started 2019-06-03.160945
; Job done 2019-06-03.160946
; Seconds 0.55
; user 0.55
; system 0.06
; cuser 0.42
; csystem 0.05




