RegulonDB
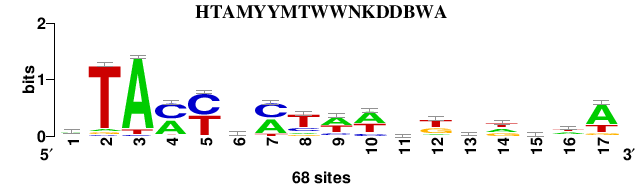
NarL matrix and aligment
matrix-quality result
Command: matrix-quality -v 1 -ms ./data/Matrices_NR/NarL/NarL.EcolK12_2nt_upstream.17.meme
Figures
Matrix logo

Decreasing cumulative distributions (dCDF)

Decreasing cumulative distributions (dCDF), logarithmic Y axis

ROC curve (logarithmic X axis)

Matrix information
; convert-matrix -v 1 -from transfac -i /NarL.EcolK12_2nt_upstream.17.meme_quality_logo
; Input files
; input /NarL.EcolK12_2nt_upstream.17.meme_quality_matrix.tf
; prior /NarL.EcolK12_2nt_upstream.17.meme_quality2nt_upstream-noorf_Escherichia_coli_GCF_000005845.2_ASM584v2-ovlp-1str.freq.gz_inclusive.tab
; Input format transfac
; Output files
; output /NarL.EcolK12_2nt_upstream.17.meme_quality_matrix_info.txt
; Output format tab
; pseudo-weight 1
; Background model
; Strand undef
; Background pseudo-frequency 0.01
; Residue probabilities
; a 0.29114
; c 0.20781
; g 0.20402
; t 0.29702
A 26 3 61 30 1 13 30 7 30 33 14 7 15 22 12 24 43
C 16 2 2 33 34 27 31 13 10 9 18 5 10 2 15 11 2
G 8 3 0 2 1 13 0 7 2 1 16 24 24 20 25 7 6
T 18 60 5 3 32 15 7 41 26 25 20 32 19 24 16 26 17
//
A 0.4 0.0 0.9 0.4 0.0 0.2 0.4 0.1 0.4 0.5 0.2 0.1 0.2 0.3 0.2 0.4 0.6
C 0.2 0.0 0.0 0.5 0.5 0.4 0.5 0.2 0.1 0.1 0.3 0.1 0.1 0.0 0.2 0.2 0.0
G 0.1 0.0 0.0 0.0 0.0 0.2 0.0 0.1 0.0 0.0 0.2 0.4 0.4 0.3 0.4 0.1 0.1
T 0.3 0.9 0.1 0.0 0.5 0.2 0.1 0.6 0.4 0.4 0.3 0.5 0.3 0.4 0.2 0.4 0.3
//
A 0.3 -1.8 1.1 0.4 -2.7 -0.4 0.4 -1.0 0.4 0.5 -0.3 -1.0 -0.3 0.1 -0.5 0.2 0.8
C 0.1 -1.9 -1.9 0.8 0.9 0.6 0.8 -0.1 -0.3 -0.4 0.2 -1.0 -0.3 -1.9 0.1 -0.2 -1.9
G -0.5 -1.5 -4.2 -1.9 -2.5 -0.1 -4.2 -0.7 -1.9 -2.5 0.1 0.5 0.5 0.4 0.6 -0.7 -0.8
T -0.1 1.1 -1.4 -1.8 0.5 -0.3 -1.0 0.7 0.2 0.2 -0.0 0.5 -0.1 0.2 -0.2 0.2 -0.2
//
A 0.1 -0.1 1.0 0.2 -0.1 -0.1 0.2 -0.1 0.2 0.2 -0.1 -0.1 -0.1 0.0 -0.1 0.1 0.5
C 0.0 -0.1 -0.1 0.4 0.4 0.3 0.4 -0.0 -0.1 -0.1 0.1 -0.1 -0.1 -0.1 0.0 -0.0 -0.1
G -0.1 -0.1 -0.0 -0.1 -0.0 -0.0 -0.0 -0.1 -0.1 -0.0 0.0 0.2 0.2 0.1 0.2 -0.1 -0.1
T -0.0 0.9 -0.1 -0.1 0.2 -0.1 -0.1 0.4 0.1 0.1 -0.0 0.2 -0.0 0.1 -0.1 0.1 -0.0
//
; Sites 68
>site_0
ATACCCATTAAGGAGTA
>site_1
ATACCCCGATCGGGGTA
>site_2
ATAACCCTTACGTAGCA
>site_3
ATACCCCTTTAGGATAA
>site_4
CTACTCATTAATGGGCA
>site_5
CTACCGCTATTGAGGTA
>site_6
TTACTCACTTCTGGGTA
>site_7
TTACCCCTTAATTATTA
>site_8
CTAACTCTAAAGTGGTA
>site_9
CTAATCCTTTATGGTTA
>site_10
ATACCTACATTGGAGTA
>site_11
ATAATGCTTTGTTAGTA
>site_12
TTAACCCAAAATGGGTA
>site_13
GTACCTATAAAGGAGCA
>site_14
GTACTCACTATGGGTAA
>site_15
ATAACGATCAATGTTAA
>site_16
CTACCCTTACCGATGTA
>site_17
ATACCCATACCCGGAAA
>site_18
CTACCTATCTCTTTGAT
>site_19
ATACCATTTATTGGTTA
>site_20
TTACTCCTTATTTGCCG
>site_21
TTACTCCGTATTTGCAT
>site_22
ATACTACTTTCGAGTGA
>site_23
GTAATTCAAAGGGAGTA
>site_24
ATACCTCACAGTGAATA
>site_25
CTAACGAATAGTAAGTA
>site_26
GTAATTATAAGGTTAAA
>site_27
TTACCGATATTTAACCT
>site_28
TTAATGATTAGTCTGAG
>site_29
ATACTCCTTAATACCCA
>site_30
TTACTCCTCACTTACAC
>site_31
ATAATCCGCCTTCGCAA
>site_32
ATAACCACACTGTGAAT
>site_33
TTAATTACATCCGTCAA
>site_34
TTACCACACATTTATTA
>site_35
CTAACCACACGGCAAAT
>site_36
CTACCTCGATGGTTTAG
>site_37
ATACCACCAAAAAGATA
>site_38
ATAACTTTATCTATCAT
>site_39
CTACCGCCAGAGAGGTA
>site_40
ATTATGATAAGTTGGAT
>site_41
ATAATACGTATGTTTGA
>site_42
TAAATGCTAATGGTGTT
>site_43
GTAACAATATCGATCGT
>site_44
GTATTCCCCATGGGTAA
>site_45
TTAATCAATTGTAAGTG
>site_46
ATAATCACATTATTTTT
>site_47
ATCCCCATCACTCTTGA
>site_48
TTACTCATTCAATTTTT
>site_49
TAAATAACATGTGTGAA
>site_50
ACAATACTTTCTGGCGA
>site_51
CAACTCATTTGTTAATT
>site_52
CTACGCTTTATTAACAA
>site_53
ATTACACTAATGCTTCT
>site_54
TTATCCATCAGACTATA
>site_55
GTTCTGTTTTGTATGAA
>site_56
ATATCAATTTCTCATCT
>site_57
TTACTTCGGCCGGTACA
>site_58
ATAATTAACAGCCACCA
>site_59
CGAATACTATGGGCAAA
>site_60
TTACTACCAAAAATAGT
>site_61
TTTACTTTATTTTTCAT
>site_62
CGAACGCTATTCCACTG
>site_63
ATTATTTTATTAATGCA
>site_64
ATAGTGAGTACAGTGAC
>site_65
GCACCACTGCCTGACAG
>site_66
CTCGCGATAATCCTATT
>site_67
CGAAATACTCCTTAGGA
;
; Matrix parameters
; Number of sites 68
; Columns 17
; Rows 4
; Alphabet A|C|G|T
; Prior A:0.291144263562772|C:0.207810681831126|G:0.204024081631615|T:0.297020972974487|a:0.291144263562772|c:0.207810681831126|g:0.204024081631615|t:0.297020972974487
; program transfac
; matrix.nb 1
; accession HTAMYYMTWWNKDDBWA
; AC HTAMYYMTWWNKDDBWA
; id HTAMYYMTWWNKDDBWA
; name HTAMYYMTWWNKDDBWA
; description mTAmycmtwwskgdswa
; statistical_basis 68 sequences
; sites 68
; nb_sites 68
; min.prior 0.204024
; alphabet.size 4
; max.bits 2
; total.information 4.57055
; information.per.column 0.268856
; max.possible.info.per.col 1.58952
; consensus.strict aTAcccctaacggggta
; consensus.strict.rc TACCCCGTTAGGGGTAT
; consensus.IUPAC mTAmycmtwwskgdswa
; consensus.IUPAC.rc TWSHCMSWWAKGRKTAK
; consensus.regexp [ac]TA[ac][ct]c[ac]t[at][at][cg][gt]g[agt][cg][at]a
; consensus.regexp.rc T[AT][CG][ACT]C[AC][CG][AT][AT]A[GT]G[AG][GT]TA[GT]
; residues.content.crude.freq a:0.3209|c:0.2076|g:0.1375|t:0.3339
; G+C.content.crude.freq 0.345156
; residues.content.corrected.freq a:0.3205|c:0.2076|g:0.1385|t:0.3334
; G+C.content.corrected.freq 0.346122
; min(P(S|M)) 9.09175e-24
; max(P(S|M)) 2.37009e-06
; proba_range 2.37009e-06
; Wmin -27.9
; Wmax 10.5
; Wrange 38.4
; logo file:/NarL.EcolK12_2nt_upstream.17.meme_quality_logo_m1.png
; logo file:/NarL.EcolK12_2nt_upstream.17.meme_quality_logo_m1_rc.png
;
; Host name sinik
; Job started 2019-06-03.161029
; Job done 2019-06-03.161030
; Seconds 0.56
; user 0.56
; system 0.05
; cuser 0.38
; csystem 0.05




