Get Started Using RegulonDB
Get Started
With RegulonDB, you can get mechanistic information about operon organization and their decomposition into transcription units (TUs), promoters and their sigma type, genes and their ribosome binding sites, terminators, binding sites of specific transcriptional regulators (TRs), as well as their organization into regulatory phrases, active and inactive conformations of TRs and simple and complex regulons.
To use RegulonDB, you need the following elements:
- An Internet connection. A broadband Internet connection
improves
performance, but it is not necessary because RegulonDB transfers only
small amounts of data (such as display data and keyboard data) to your computer.
- The menu and tools of RegulonDB use JavaScript, for this
reason, we recommend to use the latest version of Internet Explorer, Firefox, Safari or a similar browser.
This how-to article assumes your computer is part of a corporate network in which remote connections are permitted. If you are unsure, ask your system administrator.
Start a Search
Once you have connected to RegulonDB home
page, you are ready to start
searching.
- In the main page, go to the search menu
- The
Search menu has eight options to search, choose one.
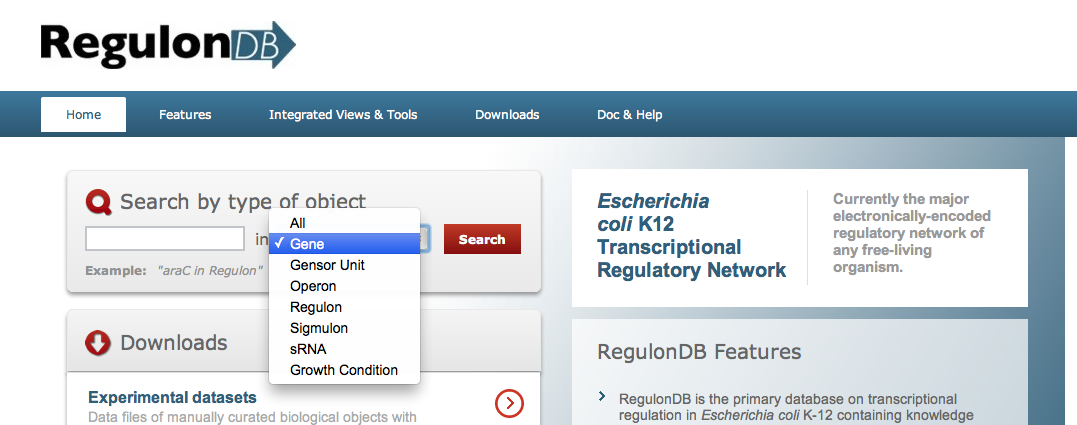
- In Gene
search, you can get information related to the gene, gene product,
Shine Dalgarno sequence, gene regulators, plus the operon and all
transcription units the gene belongs to. You can also see a graphic
display containing all objects located within the context region,
including promoters, binding sites and terminators. This may also
include sites with no effect on regulation of the query-gene.
- Operon
search. A common definition of operon is: a set of contiguous
genes
co-transcribed in a given condition. For database purposes we define
two terms in RegulonDB: operon and transcription unit, where operon is a set of one or more
overlapping transcription units which share
genes. In RegulonDB, one gene cannot belong to more than one operon. A transcription unit is a set of one
or more genes transcribed from a single promoter. A TU may also include
regulatory protein binding sites affecting this promoter and a
terminator.
From this search, you can get information related to the operon, all the transcriptions units -TU- belonging to the operon, plus the regulation for each TU. The Operon graph is displayed with all the regulatory elements located within the context region. The complete set of known transcriptions units are displayed below the operon with their detailed regulatory information. - In the Regulon
search, you can get simple and complex regulon information. The classic definition of a regulon is a
group of genes regulated by one and only one transcription factor (Maas
WK, 1964, PMID:14168690).
We call this a simple regulon.
A complex
regulon is
defined as a group of genes regulated by several transcriptional
regulators, where each regulator has the same effect on all the genes
of the regulon.
All the binding sites and promoters grouped by function are displayed.
- Growth
Condition search. At the present, this data can be accessed from
download option, we are working to generate specific
search and navigation tools.