| If I have a gene, how can I get all what is known about its regulation and operon organization of melA?
|
|---|
Frequent Navigation Scenarios
| If I have a gene, how can I get all what is known about its regulation and operon organization of melA?
|
|---|
|
|
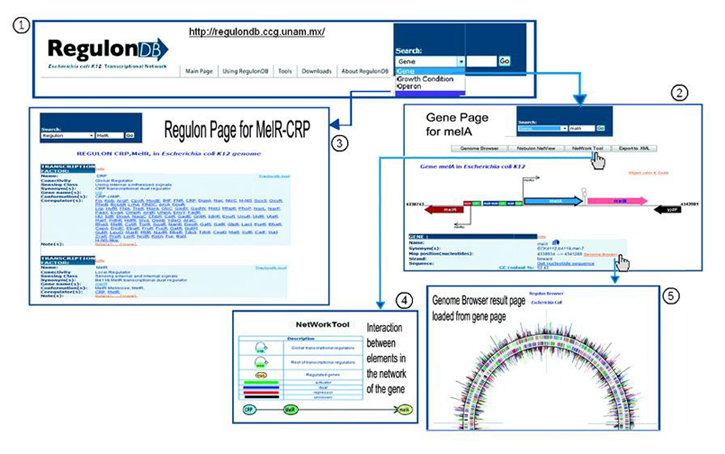
1. Go to RegulonDB main page, use the search box and write melA. Then, choose the option "gene" and click on go. You can also choose the option "Regulon" to go to "Regulon page".
2. The next page will display the graphical context and text information of the gene. You can also find its product, the operon it belongs to and a summary of its regulation. Use the link "Display regulation" to go to "Operon page", or click on Network Tools to see the graphical display of the regulation. 3. On the regulon page, you can see the details of the regulon melA belongs to, the description of the transcription factors, the DNA binding sites they bound, including the details of the promoter, function and central distance to the transcriptional start site. 4. The graphical display of the regulation shows the elements that regulate the gene, as well as the elements that are regulated by the gene itself, including autoregulation. For instance, it is possible to observe that melA is activated by CRP and regulated on a dual form by MelR. 5. The Genome Browser shows the localization of melA in the genome. |
| If we have a set of genes coming from a ChIP-chip experiment with LexA, how can we discover the transcription factor DNA-binding sites (TFDBSs) common to a regulon obtained from RegulonDB? |
|
|---|
|
|
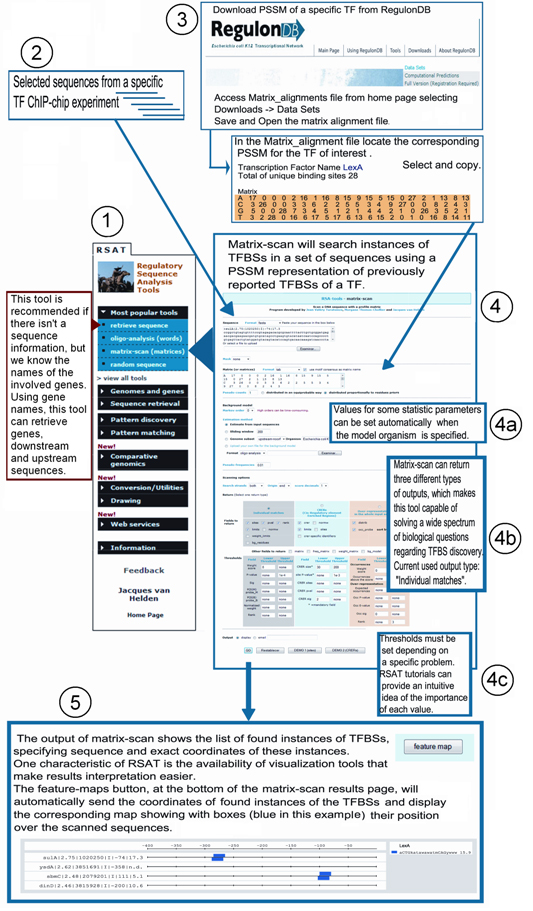
1. Go to RSA Tools main page from RegulonDB - Tools - RSA Tools. This tool is recommended if there's a lack of sequence information but names of involved genes are known. Using gene names, this tool can retreive genes as well as downstream and upstream sequences. In this page, select the option of "Escherichia coli K12" organism, and specify the gene name "LexA".
|
| How can I have acces to Textpresso from RegulonDB?
|
|---|
|
|
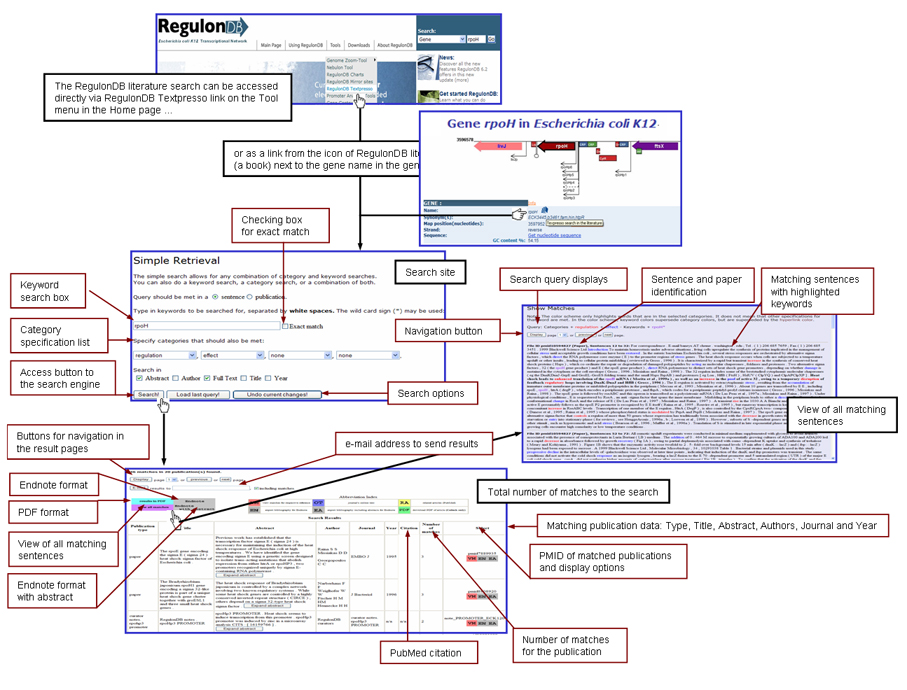
RegulonDB uses the text-mining tool TextPresso for accessing to specific gene regulation literature and full papers. The RegulonDB literature search can be accessed directly via the link Textpresso in the “Tools” option on the menu of the RegulonDB Home page or as a link on the RegulonDB Literature (book) icon, which is next to the gene name in the gene page. After clicking on this link, the RegulonDB literature search interface is displayed. For formulating a query you will first need to choose the keywords and categories involved with the information you are interested on. After initiating a search by clicking on the "Search!" button, you will get the results in the Simple Retrieval page with the total of matches to your search. The page will display the number of matches in according with the user search parameters. All publications that contain one or more hits will be shown; the search summary page has diverse output format options. In this page you will find a view sentences button (vm), which contains a link where you will be able to visualize the current sentences of the publication where matches to the query were identified. The matching sentences are presented with the search parameters highligthed. |
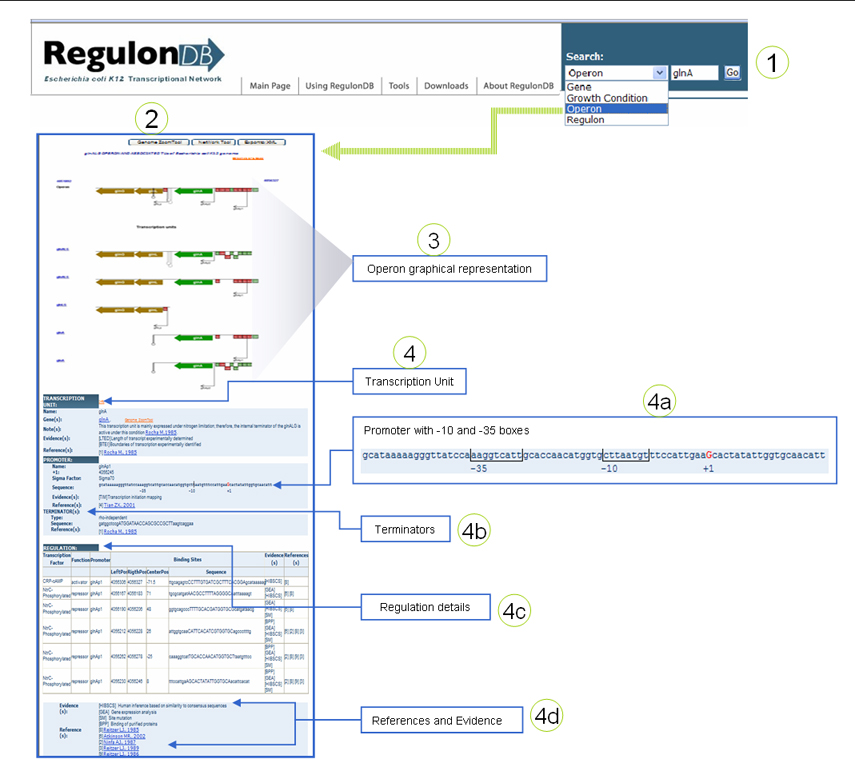
| I have the name of an operon and I want to know everything about it.
|
|---|
|
|
|